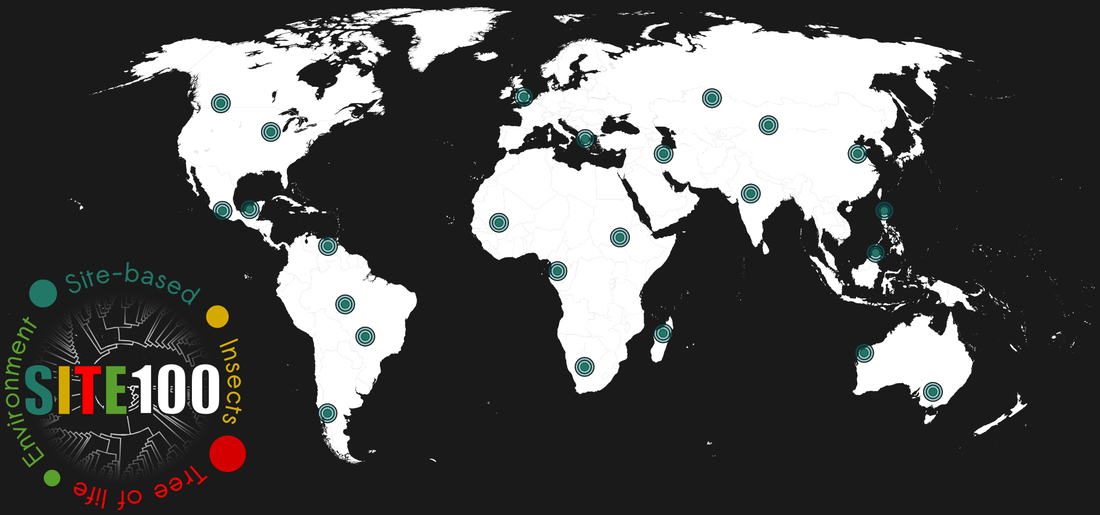
Linking site-based and lineage-based analyses
The set of some 100 sites are selected to represent a significant subsample of the existing insect diversity on Earth, covering different biogeographic regions around the globe on a coarse grid. Full or partial mitochondrial genomes (and possibly other markers) place each species on the deep-level phylogenetic tree, while the tree provides a phylogenetic fingerprint of each site.
How the data may be used – revealing biodiversity patterns
Mitogenome data from many local sites provide a unified image of global species and phylogenetic diversity. They can be used to analyse the structure of biodiversity both on temporal and spatial scales. Individual sites may be viewed as islands within the continental landscape that each hold a certain number of ‘endemic’ species. The more densely the grid of sites is sampled, the fewer species will be unique to any site, and this is even more true at the level of clades, which have wider distributions than single species.
A spatial and evolutionary continuum
Distribution data from site-based analysis and evolutionary data from the phylogenetic tree work together to provide the two major dimensions of biodiversity patterns, space and time. If lineages diverge through time while their ranges move gradually, the evolutionary depth of lineage separation is expected to mirror the spatial divergence. Thus, certain levels of the evolutionary hierarchy define the extent of biogeographic regions and the relative age of endemic communities. The SITE100 database can test these hypotheses and assess them for particular lineages and geographic regions, for a map of evolutionary divergence and spatial uniqueness of biological diversity.
The set of some 100 sites are selected to represent a significant subsample of the existing insect diversity on Earth, covering different biogeographic regions around the globe on a coarse grid. Full or partial mitochondrial genomes (and possibly other markers) place each species on the deep-level phylogenetic tree, while the tree provides a phylogenetic fingerprint of each site.
How the data may be used – revealing biodiversity patterns
Mitogenome data from many local sites provide a unified image of global species and phylogenetic diversity. They can be used to analyse the structure of biodiversity both on temporal and spatial scales. Individual sites may be viewed as islands within the continental landscape that each hold a certain number of ‘endemic’ species. The more densely the grid of sites is sampled, the fewer species will be unique to any site, and this is even more true at the level of clades, which have wider distributions than single species.
A spatial and evolutionary continuum
Distribution data from site-based analysis and evolutionary data from the phylogenetic tree work together to provide the two major dimensions of biodiversity patterns, space and time. If lineages diverge through time while their ranges move gradually, the evolutionary depth of lineage separation is expected to mirror the spatial divergence. Thus, certain levels of the evolutionary hierarchy define the extent of biogeographic regions and the relative age of endemic communities. The SITE100 database can test these hypotheses and assess them for particular lineages and geographic regions, for a map of evolutionary divergence and spatial uniqueness of biological diversity.
What criteria for selecting a site?
We attempt to obtain a sample that serves to describe the taxonomic diversity in (mostly) pristine sites, as a representation of the regional biodiversity. We are targeting primary forests or largely unmodified areas wherever possible. That will include some classical tropical and temperate field sites, in particular those with long time series or ongoing research. The main aim is the generation of a reference library of local species and their phylogenetic placement based on mitochondrial genomes. In addition, larger trap catches will be analysed with PCR-based methods to assess the total diversity, and the variation between traps at the local scale.
How to define a ‘site’?
A ‘site’ refers to a limited geographic region of interest to study global insect diversity. This may be a single nature reserve of limited extent, or a distinct biogeographic or political area harbouring a discrete fauna, such as ‘Southeastern England’. Whatever the overall scope of a particular study, we attempt to set up local plots of some 1 ha that are sampled in a standardised way if at all possible (see Methodology -> Field Protocols). The aim of SITE100 inventories is to enable more detailed local studies, which may be carried out with other methods, such as metabarcoding. The mitogenome data and higher-level tree link these local studies to the global evolutionary diversity.
How many species should be obtained from each site?
Sampling is conducted in a semi-standardised way using a set of traps (see Field Protocols), and the success of these techniques and the local diversity determine how many species are obtained. But how many species should we aim for at a sampling site to make a solid contribution to the global database? In the case of the Coleoptera, we aim to obtain ~1000 species at each site. Given 100 sites, a high proportion (possibly 10%) of the total species diversity thought to exist will be obtained, and this proportion is higher for each of level of the phylogenetic hierarchy. The proportion of total species diversity captured at each target site ultimately depends on the degree to which the local species and higher lineages are endemic, and the spatial scale at which they are undergoing turnover (beta diversity).
Why 100 sites?
Analysing the spatial scale of turnover, at different hierarchical levels, is the primary aim of SITE100. The more sites are being analysed, the greater is the resolution at which these patterns can be revealed. However, our current understanding of species and phylogenetic distributions is too poor, and the true diversity presumably is too unpredictable, to decide on a precise number of sites needed. 100 sites are a compromise between feasibility and the level of spatial and phylogenetic resolution needed to understand global diversity patterns.
What will a local investigators gain from the data?
SITE100 brings together different local studies under a single umbrella. People working at a site may be interested in various factors relevant to particular groups or particular areas, such as studies of spatial patterns and habitat differences at local scales, or the effects of disturbance gradients or environmental change over time. These specific topics will not be directly addressed by the SITE100 study. However, by putting the local specimens in the context of the wider data, the local studies will help to characterise the local communities better. For example, the local fauna may turn out to be highly endemic at the phylogenetic level, and thus of great conservation value. Or it may consist of lineages that have a tendency to be widespread, e.g. in disturbed area, giving an idea of the homogenisation globally. The SITE100 project primarily represents the global taxonomic and phylogenetic context. In addition, using similar sampling and sequencing protocols, local diversity can be assessed against similar studies conducted elsewhere, to extract common factors.
We attempt to obtain a sample that serves to describe the taxonomic diversity in (mostly) pristine sites, as a representation of the regional biodiversity. We are targeting primary forests or largely unmodified areas wherever possible. That will include some classical tropical and temperate field sites, in particular those with long time series or ongoing research. The main aim is the generation of a reference library of local species and their phylogenetic placement based on mitochondrial genomes. In addition, larger trap catches will be analysed with PCR-based methods to assess the total diversity, and the variation between traps at the local scale.
How to define a ‘site’?
A ‘site’ refers to a limited geographic region of interest to study global insect diversity. This may be a single nature reserve of limited extent, or a distinct biogeographic or political area harbouring a discrete fauna, such as ‘Southeastern England’. Whatever the overall scope of a particular study, we attempt to set up local plots of some 1 ha that are sampled in a standardised way if at all possible (see Methodology -> Field Protocols). The aim of SITE100 inventories is to enable more detailed local studies, which may be carried out with other methods, such as metabarcoding. The mitogenome data and higher-level tree link these local studies to the global evolutionary diversity.
How many species should be obtained from each site?
Sampling is conducted in a semi-standardised way using a set of traps (see Field Protocols), and the success of these techniques and the local diversity determine how many species are obtained. But how many species should we aim for at a sampling site to make a solid contribution to the global database? In the case of the Coleoptera, we aim to obtain ~1000 species at each site. Given 100 sites, a high proportion (possibly 10%) of the total species diversity thought to exist will be obtained, and this proportion is higher for each of level of the phylogenetic hierarchy. The proportion of total species diversity captured at each target site ultimately depends on the degree to which the local species and higher lineages are endemic, and the spatial scale at which they are undergoing turnover (beta diversity).
Why 100 sites?
Analysing the spatial scale of turnover, at different hierarchical levels, is the primary aim of SITE100. The more sites are being analysed, the greater is the resolution at which these patterns can be revealed. However, our current understanding of species and phylogenetic distributions is too poor, and the true diversity presumably is too unpredictable, to decide on a precise number of sites needed. 100 sites are a compromise between feasibility and the level of spatial and phylogenetic resolution needed to understand global diversity patterns.
What will a local investigators gain from the data?
SITE100 brings together different local studies under a single umbrella. People working at a site may be interested in various factors relevant to particular groups or particular areas, such as studies of spatial patterns and habitat differences at local scales, or the effects of disturbance gradients or environmental change over time. These specific topics will not be directly addressed by the SITE100 study. However, by putting the local specimens in the context of the wider data, the local studies will help to characterise the local communities better. For example, the local fauna may turn out to be highly endemic at the phylogenetic level, and thus of great conservation value. Or it may consist of lineages that have a tendency to be widespread, e.g. in disturbed area, giving an idea of the homogenisation globally. The SITE100 project primarily represents the global taxonomic and phylogenetic context. In addition, using similar sampling and sequencing protocols, local diversity can be assessed against similar studies conducted elsewhere, to extract common factors.