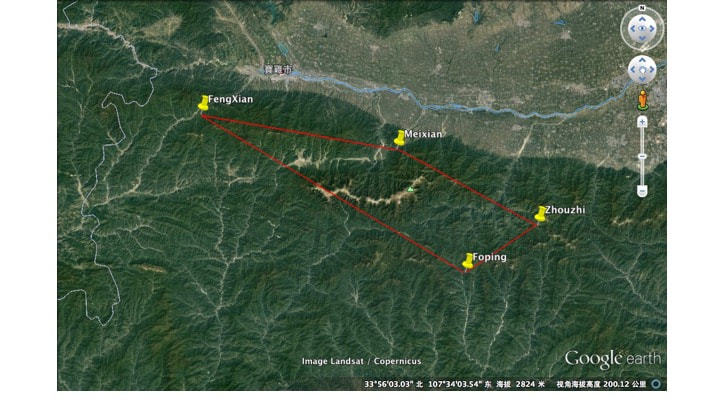
Qingling Mountains, China
The Qinling Mountains, Shaanxi Province, are best known for the Panda preservation programs but also harbour a great diversity of insects, which have a long history of study. An extensive sampling campaign was conducted by the Chinese Academy of Sciences under the direction of Dr Nie Ruie in the summer of 2016.
Site name:
Project lead: RuiE Nie, Chinese Academy of Sciences
Status of project:
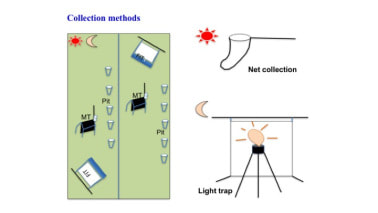
Sampling at the four main sites consisted of 10 plots each, sampled with FIT, Malaise traps, 10 pitfall traps, and beating of vegetation (three weeks)
(a) Number of DNA grade specimens (Coleoptera only)
Feng Xian: 12462
Fo Ping: 7756
Mei Xian: 9918
Zhouzhi: 9614
(b) Reference specimen set:
1248 specimens each representing separate ‘morphospecies’ (DNA extraction and images)
(c) DNA extractions: number of extractions, single or bulk
Bulk samples 1292 tubes of pools of either 50 or 25 specimens each
(d) DNA sequencing: genes sequenced, sequencing methodology
COI barcode/ metabarcode and shotgun (pooled morphospecies)
Sequencing methodology: Illumina sequencing platform_Hiseq 2500
(e) metabarcoding: marker used, number of samples
COI: primers: F:CCNGAYATRGCNTTYCCNCG&R: TANACYTCNGGRTGNCCRAARAAYCA with different tags
(f) Sequence data: (Genbank accession xx-xx)
Not available
- FengXian (1512 – 1785 m a.s.l.; 34.21 – 34.18 N; 106.902 – 106.915 E;
- Foping (1468 – 1538 m a.s.l; 33°40’ N; 107°58’ E;
- MeiXian (1118-1309 m a.s.l., 34°4’52N;107°42′ E,
- Zhouzhi (1350- 1564 m, 33°49′ N;108°15′ E
Project lead: RuiE Nie, Chinese Academy of Sciences
Status of project:
Sampling at the four main sites consisted of 10 plots each, sampled with FIT, Malaise traps, 10 pitfall traps, and beating of vegetation (three weeks)
(a) Number of DNA grade specimens (Coleoptera only)
Feng Xian: 12462
Fo Ping: 7756
Mei Xian: 9918
Zhouzhi: 9614
(b) Reference specimen set:
1248 specimens each representing separate ‘morphospecies’ (DNA extraction and images)
(c) DNA extractions: number of extractions, single or bulk
Bulk samples 1292 tubes of pools of either 50 or 25 specimens each
(d) DNA sequencing: genes sequenced, sequencing methodology
COI barcode/ metabarcode and shotgun (pooled morphospecies)
Sequencing methodology: Illumina sequencing platform_Hiseq 2500
(e) metabarcoding: marker used, number of samples
COI: primers: F:CCNGAYATRGCNTTYCCNCG&R: TANACYTCNGGRTGNCCRAARAAYCA with different tags
(f) Sequence data: (Genbank accession xx-xx)
Not available