The SITE-100 project is an ambitious effort towards documenting the species diversity and evolutionary history of insects on Earth, using genomic sequencing and studying selected sites around the world. Arthropods can be sampled with standard trapping methods (Malaise, flight-interception, pitfall) which usually produce thousands of specimens, but these complex mixtures (‘biodiversity soup’) are largely intractable to conventional taxonomy.
Metagenomics and metabarcoding procedures are used for the characterisation of these specimen mixtures, to to place the local diversity into species-rich phylogenetic tree representing the global diversity. Short ‘barcode’ and 'metabarcode' sequences obtained from PCR amplification of bulk samples are identified by phylogenetic placement. Linking genomic sequencing with field-based identification techniques supports the much needed efforts for the long-term preservation of insect diversity locally and globally.
Metagenomics and metabarcoding procedures are used for the characterisation of these specimen mixtures, to to place the local diversity into species-rich phylogenetic tree representing the global diversity. Short ‘barcode’ and 'metabarcode' sequences obtained from PCR amplification of bulk samples are identified by phylogenetic placement. Linking genomic sequencing with field-based identification techniques supports the much needed efforts for the long-term preservation of insect diversity locally and globally.
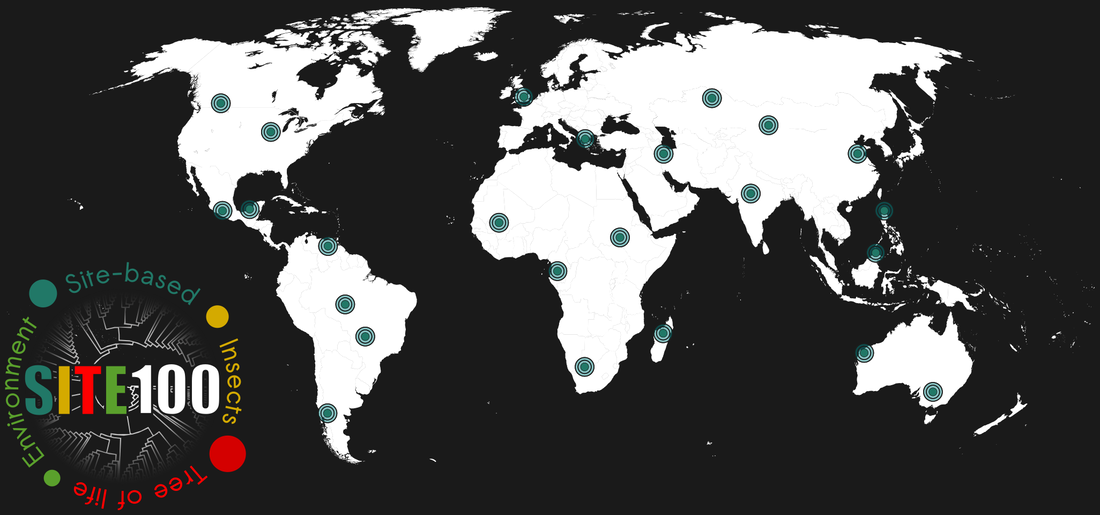
Existing insect taxonomy has left up to 90% of species yet to be described. SITE-100 uses intensive surveys at local sites (‘observatories’) to increase the sampling of global diversity, and generates DNA data that connect local sites to the global effort of taxonomy and species discovery. Our sites include classical locations of biodiversity studies and new sites that form part of local research and survey work. Site-based approaches are maximising the number of collected species while minimizing the logistics of field work.
As biodiversity studies are increasingly using DNA data, the SITE-100 project provides a platform for connecting these local studies through a largely uniform set of sequence information. Biodiversity genomics needs to balance the desire for powerful phylogenetic markers and the feasibility of analysing large numbers of specimens. With current technology, mitochondrial genomes are the most suitable data to accomplish robust phylogenetic trees and high specimen throughput.
Local studies in SITE-100 network use similar field sampling protocols, DNA sequencing methods and bioinformatics procedures. Data from different sites work synergistically and provide an increasingly complete picture of species diversity and distribution. When combined among sites, and together with external public sequence data, they yield a detailed phylogenetic tree of the insects. In turn, the local studies benefit from having available a global database of species occurrences and phylogenetic position. Ultimately, the data can be used to parameterise models of diversity across geographic and temporal scales, for a heat map of global species diversity of the Insecta and to study the effects of environmental change.
We aim to include 100 such local sites from around the world, to cover all major biogeographic regions. The focus is on species-rich, natural forest ecosystems in the tropics and temperate zone.
As biodiversity studies are increasingly using DNA data, the SITE-100 project provides a platform for connecting these local studies through a largely uniform set of sequence information. Biodiversity genomics needs to balance the desire for powerful phylogenetic markers and the feasibility of analysing large numbers of specimens. With current technology, mitochondrial genomes are the most suitable data to accomplish robust phylogenetic trees and high specimen throughput.
Local studies in SITE-100 network use similar field sampling protocols, DNA sequencing methods and bioinformatics procedures. Data from different sites work synergistically and provide an increasingly complete picture of species diversity and distribution. When combined among sites, and together with external public sequence data, they yield a detailed phylogenetic tree of the insects. In turn, the local studies benefit from having available a global database of species occurrences and phylogenetic position. Ultimately, the data can be used to parameterise models of diversity across geographic and temporal scales, for a heat map of global species diversity of the Insecta and to study the effects of environmental change.
We aim to include 100 such local sites from around the world, to cover all major biogeographic regions. The focus is on species-rich, natural forest ecosystems in the tropics and temperate zone.
100 Observatories
We envision that 100 such observatories characterised in this way are adequate to cover the ecological, biogeographic and phylogenetic diversity on Earth at sufficient resolution to provide an evolutionary and taxonomic framework for the Insecta. In this first phase of the project we will use the Coleoptera (beetles) as a pilot group.
Site-based studies have the advantage of:
Sampling
At each site, standardized protocols are used for:
We envision that 100 such observatories characterised in this way are adequate to cover the ecological, biogeographic and phylogenetic diversity on Earth at sufficient resolution to provide an evolutionary and taxonomic framework for the Insecta. In this first phase of the project we will use the Coleoptera (beetles) as a pilot group.
Site-based studies have the advantage of:
- simplified logistics, including permits
- work with local partners (academic and citizen science)
- repeat visits to conduct long-term trapping
- study seasonal and environmental change
Sampling
At each site, standardized protocols are used for:
- maximum coverage of arthropod species
- uniform spatial design
- generating DNA grade specimen
- calibrating short sampling against permanent series